2026

Structure and signaling mechanism of Helicobacter pylori transducer-like protein D.
Franco, K., DiIorio, M., Simpkin, A.J., Keegan, R.M., Kallio, K., Goers-Sweeney, E., Colbert, M., Remington, S.J., Cassidy, C.K., Kulczyk, A.W.*, Baylink, A*.
*Corresponding authors
bioRxiv, doi.org/10.64898/2026.01.16.699579 (2026).
2025
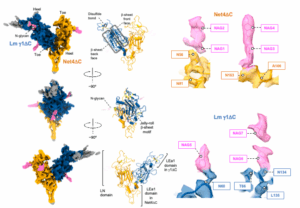
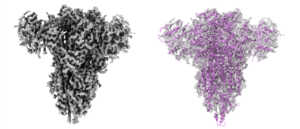
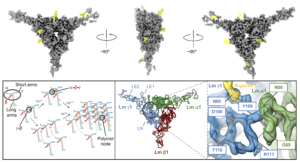
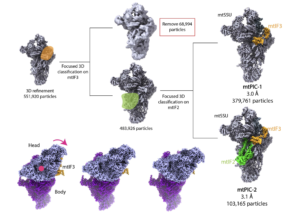
Cryo-EM reveals molecular mechanisms underlying the inhibitory effect of netrin-4 on laminin matrix formation.
Kulczyk, A.W.*, McKee, K.K., Yurchenco, P.D.
*Corresponding author
Nature Communications, 16(1), 7256-7270 (2025).
2024
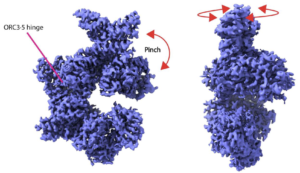
Cryo-electron microscopy studies of biomolecular structure and dynamics.
Kulczyk, A.W.
Micromachines, 15(9), 1092-1095 (2024).
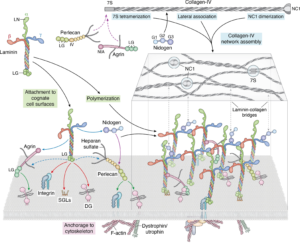
Polymerizying laminins in development, health and disease.
Yurchenco, P.D., Kulczyk, A.W.
Journal of Biological Chemistry, 300(7), 107429-107449 (2024).
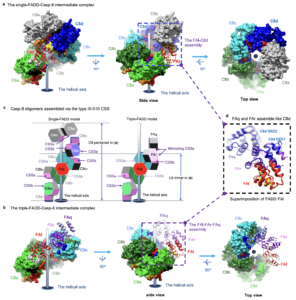
Deciphering DED assembly mechanisms in FADD-procaspase-8-cFLIP complexes regulating apoptosis.
Yang, C-Y., Lien, C-I., Tseng, Y-C., Tu, Y-F., Kulczyk, A.W., Lu, Y-C., Wang, Y-T., Su, T-W., Hsu, L-C., Lin, S-C.
Nature Communications, 15(1):3791-3809 (2024).
2023
Artificial intelligence and the analysis of cryo-EM data provide insight into the molecular mechanisms underlying LN-lamininopathies.
Kulczyk, A.W.
Scientific Reports , 13(1):17825-17840 (2023).
Novel artificial intelligence-based approaches for ab initio structure determination and atomic model building for cryo-electron microscopy.
DiIorio, M.C., Kulczyk, A.W.
Micromachines, 14(9), 1674-1700 (2023).
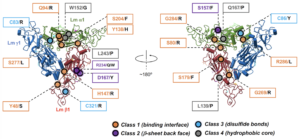
Cryo-EM reveals the molecular basis of laminin polymerization and LN-lamininopathies.
Kulczyk, A.W.*, McKee, K.K., Zhang, X., Bizukojc, I., Yu, Y.Q., Yurchenco, P.D.
*Corresponding author
Nature Communications, 14(1), 317-324 (2023).

Cryo-EM structure of Shiga toxin 2 in complex with the native ribosomal P-stalk reveals residues involved in the binding interaction.
Kulczyk, A.W.*, Sorzano, C.O.S., Grela, P., Tchorzewski, M., Tumer, N.E., Li, X-P.
*Corresponding author
Journal of Biological Chemistry, 299(1), 102795-102808 (2023).

YlaN is an iron(II) binding protein that functions to releve Fur-medieted repression of gene expression in Staphylococcus aureus.
Boyd, J.M., Esquilin-Lebron, K.M., Cambell, C.J., Kaler, K.M.R., Norambuena, J., Foley, M.E., Stephens, T.G., Rios, G., Mereddy, G., Zheng, V., Bovermann, H., Kim, J., Kulczyk, A.W., Yang, J.H., Greco, T.M., Cristea, I.M., Carabetta, V.J., Beavers, V.N., Bhattacharya, D., Skaar, E.P., Parker, D., Carroll, R.K., Stemmler, T.L.
bioRxiv, doi.org/10.1101/2023.10.03.560778 (2023).
2022
Exploring the structural variability of dynamic biological complexes by single-particle cryo-electron microscopy.
DiIorio, M.C., Kulczyk, A.W.
Micromachines, 14(1), 118-142 (2022).
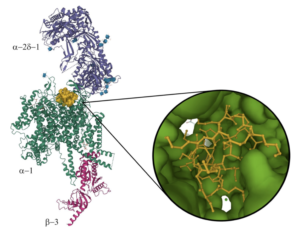
Electron microscopy holdings of the Protein Data Bank: the impact of the resolution revolution, new validation tools, and implications for the future.
Burley, S., Berman, H.M., Chiu, W., Dai, W., Flatt, J.W., Hudson, B.P., Kaelber, J.T., Khare, S.D., Kulczyk, A.W., Lawson, C.L., Pintilie, G.D., Sali, A., Vallat, B., Westbrook, J.D., Young, J.Y., Zardecki, C.
Biophysical Reviews, 14(6), 1281-1301 (2022).

You cannot oxidize what you cannot reach: Oxidative susceptibility of buried methionine residues.
Kulczyk, A.W., Leustek, T.
Journal of Biological Chemistry, 298(5), 101973-101974 (2022).

A robust single-particle cryo-electron microscopy (cryo-EM) processing workflow with cryoSPARC, Relion, and Scipion.
DiIorio, M.C., Kulczyk, A.W.
Journal of Visual Experiments, 179, doi: 10.3791/63387 (2022).
2018
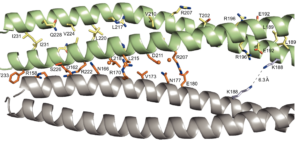
A crystal structure of coil 1B vimentin in the filamentous form provides a model of a high-order assembly of a vimentin filament.
Pang, A.H., Obiero, J.M., Kulczyk, A.W., Sviripa, V.M., Tsodikov, O.V.
FEBS Journal, 10, 1-12 (2018).
2017

Cryo-EM structure of the replisome reveals multiple interactions coordinating DNA synthesis.
Kulczyk, A.W.*, Moeller, A., Meyer P., Sliz, P., Richardson, C.C.*
*Corresponding authors
Proceedings of the National Academy of Sciences of the USA, 114, 1848-1856 (2017).
2016
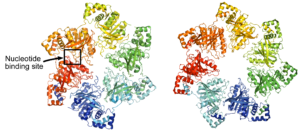
The Replication System of Bacteriophage T7.
Kulczyk, A.W., Richardson, C.C.
The Enzymes, 39, 89-136 (2016).
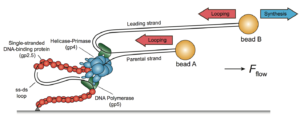
Simultaneous real-time imaging of leading- and lagging-strand synthesis reveals the coordination dynamics of single replisomes.
Duderstadt, K.E., Geertsema, H.J., Stratmann, S.A., Punter, C.M., Kulczyk, A.W., Richardson, C.C., van Oijen, A.M.
Molecular Cell, 64, 1035-1047 (2016).
Structural and functional probing of PorZ, an essential bacterial-surface component of the type-IX secretion system of human oral-microbiomic P. gingivalis.
Lasica A.M., Goulas T., Mizgalska D., Zhou X., de Diego I., Ksiazek M., Madej M., Guo Y., Guevara T., Nowak M., Potempa B., Goel A., Sztukowska M., Prabhakar A.T., Bzowska M., Widziolek M., Thøgersen I.B., Enghild J.J., Simonian M., Kulczyk, A.W., Nguyen K.A., Potempa J., Gomis-Rüth F.X.
Scientific Reports, 6, 37708 (2016).
2014
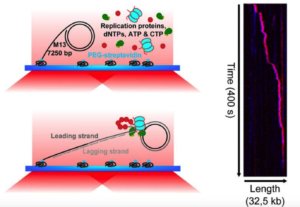
Single-molecule studies of polymerase dynamics and stoichiometry at the bacteriophage T7 replication machinery.
Geertsema, H.J., Kulczyk, A.W., Richardson, C.C., van Oijen A.
Proceedings of the National Academy of Sciences of the USA, 111, 4073-4078 (2014).
2012
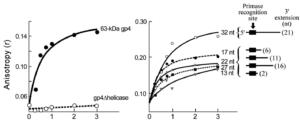
Molecular interactions in bacteriophage T7 priming complex.
Kulczyk, A.W.*, Richardson, C.C.
*Corresponding author
Proceedings of the National Academy of Sciences of the USA, 109, 9408-9413 (2012).
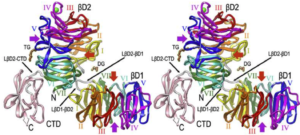
An interaction between DNA polymerase and helicase is essential for the high processivity of the replisome.
Kulczyk, A.W., Akabayov, B., Lee, S.J., Bostina M., Berkowitz S.A., Richardson C.C.
Journal of Biological Chemistry, 287(46), 39050-39060 (2012).
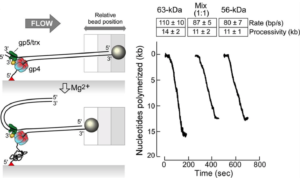
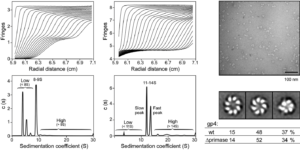
Heterohexamer of 56- and 63-kD gene 4 helicase-primase of bacteriophage T7 in DNA replication.
Zhang, H., Lee, S.J., Kulczyk, A.W., Zhu, B., Richardson, C.C.
Journal of Biological Chemistry, 287(41), 34273-34287 (2012).
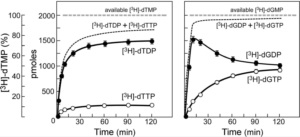
Characterization of a nucleotide kinase encoded by bacteriophage T7.
Tran, N., Amarasiriwarden, C.J., Kulczyk, A.W., Richardson, C.C., Tabor, S.
Journal of Biological Chemistry, 287 (35), 29458-29478 (2012).
2011
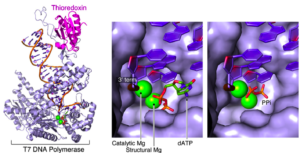
Pyrovanadolysis: a pyrophosphorolysis-like reaction mediated by pyrovanadate, Mn2+, and DNA polymerase of bacteriophage T7.
Akabayov, B., Kulczyk, A.W., Akabayov, S., McLaughlin, L., Theile, C., Beauchamp, B., Richardson, C.C.
Journal of Biological Chemistry, 286(33), 29146-29157 (2011).
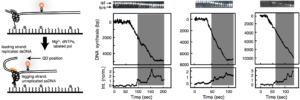
Observing polymerase exchange by simultaneous measurements of replisome structure and function at the single-molecule level.
Loparo, J., Kulczyk, A.W., Richardson, C.C., van Oijen A.
Proceedings of the National Academy of Sciences of the USA, 108, 3584-3589*.
*commentary by Shi, X., Ha, T. (2011). Seeing a molecular machine self-renew. (2011). Proceedings of the National Academy of Sciences of the USA, 108, 3459-3460 (2011).
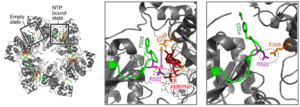
A Critical residue for coupling dTTP hydrolysis with DNA unwinding by the helicase of bacteriophage T7.
Satapathy, A.K., Kulczyk, A.W., Ghosh, S., van Oijen, A.M., Richardson, C.C.
Journal of Biological Chemistry, 286(39), 34468-34478 (2011).
2010
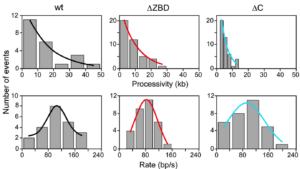
Direct observation of enzymes replicating DNA using a single-molecule DNA stretching assay.
Kulczyk, A.W., Tanner N., Loparo J., Richardson C., van Oijen A.
Journal of Visual Experiments, 37, id: 1689, doi: 10.3791/1689 (2010).
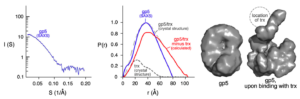
Conformational dynamics of bacteriophage T7 DNA polymerase and its processivity factor, Escherichia coli thioredoxin.
Akabayov, B., Akabayov, S.R., Lee, S.J., Tabor, S., Kulczyk, A.W., Richardson, C.C.
Proceedings of the National Academy of Sciences of the USA, 107, 15033-15038 (2010).
2008

Inadequate inhibition of host RNA polymerase restricts T7 bacteriophage growth on hosts overexpressing udk.
Qimron, U., Kulczyk, A.W., Hamdan, S.M., Tabor, S., Richardson, C.C.
Molecular Microbiology, 67, 448-457 (2008).
2004

Solution structure and DNA binding of the zinc finger domain from DNA ligase IIIa.
Kulczyk, A.W., Yang, J., Neuhaus, D.
Journal of Molecular Biology, 341, 723-738 (2004).
