Current projects
Structure determination of the human mitochondrial replisome
Nearly three hundred mutations have been identified to date in the sequence of mitochondrial replication proteins. These mutations play a causative role in development of cancer and neurodegenerative mitochondrial disorders. Due to the lack of structural data, an exact mechanism by which altered replication proteins affect replisome function is not known.
Cryo-electron microscopy (cryo-EM) has recently revolutionized structural biology (Nobel Prize in Chemistry 2017) and is particularly suitable for structural analysis of the mitochondrial replisome, because it provides capabilities for examining functional states of heterogeneous populations of large multi-protein complexes at the atomic resolution. We anticipate that the structural data will yield new insight into the mechanism by which the mutant proteins alter activities of the replisome in human mitochondria. This information may subsequently facilitate development of novel therapeutics.
We also apply cutting-edge methods including correlative light and electron microscopy (CLEM), focus ion beam-scanning electron microscopy (FIB-SEM) and cryo-electron tomography (cryo-ET) to determine the replisome structure
in situ by 3-D subtomogram averaging.
We have previously determined a structure of the 650 kDa bacteriophage T7 replisome (Kulczyk et al., 2017, PNAS; Kulczyk et al., 2012, PNAS; Kulczyk et al., 2012, JBC) homologous to the human mitochondrial replisome (Kulczyk et al., 2016, Enzymes). The cryo-EM structure defines an architecture of the entire replisome, and provides the first such structure of a functioning replication complex assembled on DNA resembling a replication fork.

Cryo-EM structure of bacteriohage T7 replisome. The structure reveals a hexameric ring of DNA helicase (in blue) to which five molecules of RNA polymerase (in orange) and two DNA polymerase-processivity factor complexes (in green/purple-yellow) bind in an asymmetric fashion. Algorithms for cryo-EM are computationally expensive. We calculated the structure using Stampede supercomputer at Texas Advanced Computing Center, the twelfth most powerful supercomputer in the world.
Structure determination of the basement membrane complexes
Basement membranes (BM) are cell-adherent extracellular matrices essential for cell signaling, differentiation, and maintenance of tissue morphology. We decipher details of the molecular interactions governing formation, plasticity, and turnover of BM by targeting structures of their key components.
One of such key components are laminins (Lm), which polymerize into a planar lattice on cell surfaces. Genetic loss or alternations in Lm subunits lead to failures of Lm polymerization manifesting in a wide spectrum of human disorders, which we collectively define as LN-lamininopathies. Interestingly, disruption of Lm polymer impedes cancer metastases and inhibits cancer angiogenesis.
We have recently determined a cryo-EM structure of the Lm polymer node, the basic repeating unit of the Lm lattice
(Kulczyk et al., 2023, Nature Commun.). The structure reveals the long-sought molecular basis of calcium-dependent formation of the Lm matrix and provides insights into polymerization defects manifesting in LN-lamininopathies
(Kulczyk, 2023, Sci. Rep.).
We also apply CLEM, FIB-SEM and cryo-ET to visualize BM
in vivo, and employ molecular dynamics simulations along with the novel artificial intelligence algorithms to study their dynamics.
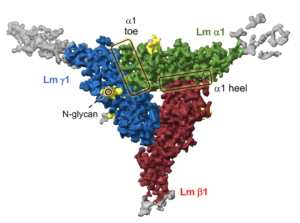
Cryo-EM structure of the Lm polymer node. The cryo-EM structure is color-coded. Lm alpha 1, beta 1 and gamma 1 subunits are shown in green, red and blue, respectively. The principal components analysis of cryo-EM data revealed the intrinsic dynamics of the Lm polymer node. Flexible parts of the Coulomb map, with no model built, are displayed in gray. In addition, the structure unveiled eight extended densities attached to its surface representing N-glycans (shown in yellow).
Single-molecule studies of the replisome dynamics
The transient nature of the protein-protein interactions that occur during synthesis of mitochondrial DNA (mtDNA) make it challenging to study the dynamics of these processes by ensemble-averaging techniques. Consequently, a number of fundamental mechanistic questions remain unanswered. For instance, what is the mechanism of mtDNA replication? To address the above and related questions we develop novel methods combining the flow-stretching of DNA and the total-internal reflection fluorescence (TIRF) microscopy; the methods allow for simultaneous monitoring of the composition and dynamics of individual fluorescently labeled replisomes during an ongoing DNA synthesis in real time. We plan to investigate mutant variants of the mitochondrial proteins that have been associated with mitochondrial disorders and cancer using these methods.
Together with colleagues from the laboratories of Charles Richardson and Antoine van Oijen at Harvard, we have applied similar methods to study bacteriophage T7 replisome (Duderstadt et al., 2016, Mol. Cell; Zhang et al., 2012, JBC; Akabayov et al., 2011, JBC; Satapathy et al., 2011, JBC; Akabayov et al., 2010, PNAS; Kulczyk et al., 2010, JoVE). For example, we have determined the mechanism of polymerase exchange on lagging- and leading-strand (Geertsema et al., 2014, PNAS and Loparo et al., 2011, PNAS, respectively).
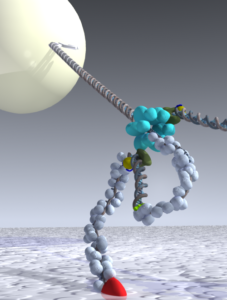
A single-molecule method to study mtDNA replication. Schematic view representing one of the single-molecule methods that we used to investigate a coordinated leading- and lagging-strand synthesis at bacteriophage T7 replisome. Please see (Loparo et al., 2011, PNAS; Kulczyk et. al., 2010, JoVE; Lee et al., Nature, 2006) for details.
Investigation of the interplay between mitochondrial DNA repair and replication
Although mitochondria were originally thought to lack DNA repair activity, recent data revealed the presence of multiple mtDNA repair pathways. Integrity of the mitochondrial genome is protected by Base Excision Repair (BER); a mechanism that functions interdependent of DNA replication. We investigate the interplay between mitochondrial short-patch BER and DNA replication using single-molecule methods that involve the hydrodynamic flow-stretching of DNA and TIRF microscopy. The DNA substrates used in the assays are modified to accommodate the damaged nucleotides, for instance 7,8-diydro-8-oxoguanine. Four enzymes are involved in a short-patch BER: DNA glycosylase, endonuclease, polymerase-gamma A (Pol-γA), and mtDNA ligase 3 (mtDL3). Interestingly, mtDL3 contains a zinc-finger (Zf) domain, which facilitates binding of Pol-γA to DNA. During doctoral studies in the MRC Laboratory of Molecular Biology at the University of Cambridge, Arek determined NMR structure of Zf from DL3 (Kulczyk et al., 2004, JMB). We currently investigate this interaction and other protein-protein interactions from the crossroads of the mitochondrial BER and DNA replication. Our goal is to expand the single-molecule studies to identify new proteins and pathways involved in repair of mtDNA.
Development of novel correlative light and electron microscopy (CLEM) techniques
We combine single-molecule techniques and cryo-EM for development of the novel methods that allow simultaneous measurements of enzymatic kinetics and structure determination. Because we have determined a structure of the T7 replisome, the phage replication complex is used as a model system. This single-molecule CLEM (sm-CLEM) will allow for systematic analysis of structural dynamics of the replisome, and in principle is applicable to the study of any other biological system.



